PCA and Biplot using Python
There are several ways to run principal component analysis (PCA) using various packages (scikit-learn, statsmodels, etc.) or even just rolling out your own through singular-value decomposition and such. Visualizing the PCA result can be done through a biplot. I was looking at an example of using prcomp and biplot in R, but it does not seem like there is a comparable plug-and-play way of generating a biplot with Python.
As it turns out, generating a biplot from the result of PCA by pcasvd of StatsModels is fairly straightforward from the rotation matrix supplied by the function. Here is a code snippet (which is also available in Gist):
#!/usr/bin/env python2.7
# -*- coding: utf-8 -*-
"""Biplot example using pcasvd from statsmodels and matplotlib.
This is an example of how a biplot (like that in R) can be produced
using pcasvd and matplotlib. Additionally, this example does k-means
clustering and color observations by which cluster they belong to.
"""
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from scipy.cluster.vq import kmeans, vq
from statsmodels.sandbox.tools.tools_pca import pcasvd
def biplot(plt, pca, labels=None, colors=None,
xpc=1, ypc=2, scale=1):
"""Generate biplot from the result of pcasvd of statsmodels.
Parameters
----------
plt : object
An existing pyplot module reference.
pca : tuple
The result from statsmodels.sandbox.tools.tools_pca.pcasvd.
labels : array_like, optional
Labels for each observation.
colors : array_like, optional
Colors for each observation.
xpc, ypc : int, optional
The principal component number for x- and y-axis. Defaults to
(xpc, ypc) = (1, 2).
scale : float
The variables are scaled by lambda ** scale, where lambda =
singular value = sqrt(eigenvalue), and the observations are
scaled by lambda ** (1 - scale). Must be in [0, 1].
Returns
-------
None.
"""
xpc, ypc = (xpc - 1, ypc - 1)
xreduced, factors, evals, evecs = pca
singvals = np.sqrt(evals)
# data
xs = factors[:, xpc] * singvals[xpc]**(1. - scale)
ys = factors[:, ypc] * singvals[ypc]**(1. - scale)
if labels is not None:
for i, (t, x, y) in enumerate(zip(labels, xs, ys)):
c = 'k' if colors is None else colors[i]
plt.text(x, y, t, color=c, ha='center', va='center')
xmin, xmax = xs.min(), xs.max()
ymin, ymax = ys.min(), ys.max()
xpad = (xmax - xmin) * 0.1
ypad = (ymax - ymin) * 0.1
plt.xlim(xmin - xpad, xmax + xpad)
plt.ylim(ymin - ypad, ymax + ypad)
else:
colors = 'k' if colors is None else colors
plt.scatter(xs, ys, c=colors, marker='.')
# variables
tvars = np.dot(np.eye(factors.shape[0], factors.shape[1]),
evecs) * singvals**scale
for i, col in enumerate(xreduced.columns.values):
x, y = tvars[i][xpc], tvars[i][ypc]
plt.arrow(0, 0, x, y, color='r',
width=0.002, head_width=0.05)
plt.text(x* 1.4, y * 1.4, col, color='r', ha='center', va='center')
plt.xlabel('PC{}'.format(xpc + 1))
plt.ylabel('PC{}'.format(ypc + 1))
def main():
"""Run a PCA on state.x77 from R and generate its biplot. Color
observations by k-means clustering."""
df = pd.io.parsers.read_csv('data/state.x77')
print df.describe()
print df.head()
columns = ['Population', 'Income', 'Illiteracy',
'Life Exp', 'Murder', 'HS Grad']
data = df[columns]
data = (data - data.mean()) / data.std()
pca = pcasvd(data, keepdim=0, demean=False)
values = data.values
centroids, _ = kmeans(values, 3)
idx, _ = vq(values, centroids)
colors = ['gby'[i] for i in idx]
plt.figure(1)
biplot(plt, pca, labels=data.index, colors=colors,
xpc=1, ypc=2)
plt.show()
if __name__ == '__main__':
main()
In addition to PCA, $k$-means clustering (three clusters) was run on the data to color the observations by how they cluster. The resulting biplot for states.x77 (which I exported and borrowed from R) looks like this:
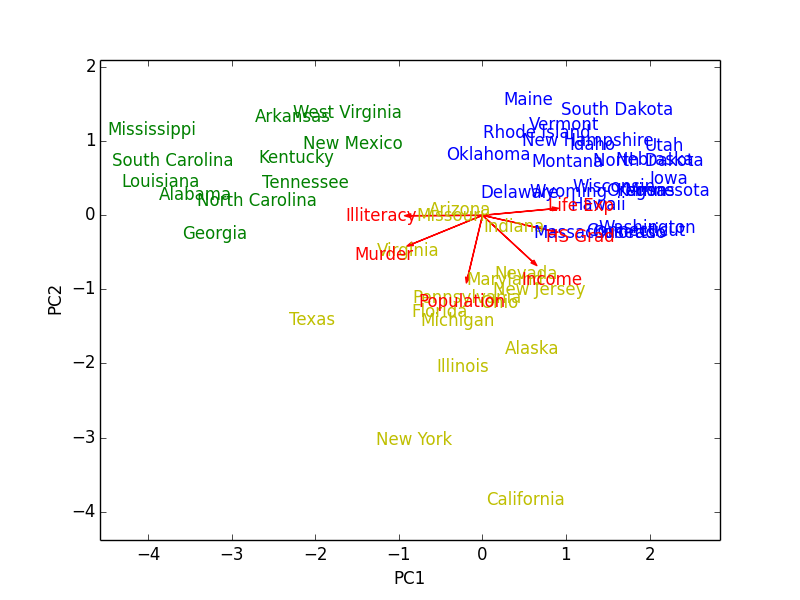
Figure 1: Biplot